Affymetrix (.cel, .cdf, .chp, .gin, .psi)
- Import fully supports the Affymetrix CEL, CDF, CHP, GIN and PSI formats.
Background & Context

-
- Affymetrix microarray data formats.
- Family of file formats used for the storage and exchange of microarray data and metainformation.
- ASCII or binary formats.
- Native formats of the MAS, GCOS and Command Console applications.
- CEL files store a raster of intensity values of individual probes.
- CHP files contain processed information about probe sets.
- CDF files describe which probes are part of which probe set.
- GIN files store the gene names associated with each probe set.
- PSI files store probe set names and the number of probe pairs in a probe set.
Import

- Import["file"] imports data from any Affymetrix CEL, CDF, CHP, GIN, or PSI file.
- Import["file",elem] imports the specified element.
- Import["file",{{elem1,elem2,…}}] imports multiple elements.
- The import format can be specified with Import["file","Affymetrix"] or Import["file",{"Affymetrix",elem,…}].
- See the following reference pages for full general information:
-
Import import from a file CloudImport import from a cloud object ImportString import from a string ImportByteArray import from a byte array
Import Elements


- General Import elements:
-
"Elements" list of elements and options available in this file "Summary" summary of the file "Rules" list of rules for all available elements - Common data representation element:
-
"Data" intensity values (CEL), processed probe set data (CHP), or probe set records (CDF, PSI, GIN) "ProbeSetNames" probe set names as a list of strings - When importing from CDF, PSI, or GIN, Import["file",{"Data",probesetname}] returns the record corresponding to the specified probe set.
- Import uses the "Data" element by default for all Affymetrix file formats.
- Common meta-information element:
-
"Header" meta-information given as list of rules - Additional CEL elements, representing meta-information about the underlying DAT image file:
-
"PixelRange" number of pixels corresponding to each probe intensity value "DataErrors" errors in intensity values "Outliers" list of coordinates of probes that are considered outliers - Additional CDF element:
-
"QCData" quality control information - Additional CHP elements:
-
"DetectionStates" detected gene expression state, given as a list of values True, False, Indeterminate "DetectionSignificances" 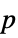 -values corresponding to each detection state
-values corresponding to each detection state"ProbePairs" number of probe pairs in each probe set "ProbePairsUsed" number of probe pairs used to infer detection states "Alleles" detected genotypes "ConfidenceValues" confidence value of each genotype detection
Examples
open allclose allBasic Examples (5)
Import and plot a raster of intensity values from a CEL file:
Import complete header information from a CEL file:
Read complete header information from a CDF file:
Import the first few probe set names:
Import data associated with a probe set name:
Import probe set names, signal data, and the detection states from a CHP file:
Read complete header information from a GIN file:
Import the first few probe set names:
Import data associated with a probe set name: