BDF (.bdf)
- Import fully supports the BDF format family.
Background & Context

-
- BDF physiological signal recordings format.
- Commonly used for the exchange and storage of medical time-series recordings.
- Application areas include electroencephalography, polysomnography, electromyography, evoked potentials, electroneurography, and electrocardiography.
- BDF is an acronym for BioSemi Data Format.
- Binary format derived from EDF.
- Stores multiple simultaneous signal channels, time-stamped annotations, and metainformation.
- Stores discontinuous time series.
- First published in 1992.
Import
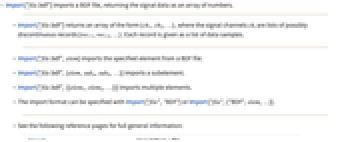
- Import["file.bdf"] imports a BDF file, returning the signal data as an array of numbers.
- Import["file.bdf"] returns an array of the form {ch1,ch2,…}, where the signal channels chi are lists of possibly discontinuous records {reci1,reci2,…}. Each record is given as a list of data samples.
- Import["file.bdf",elem] imports the specified element from a BDF file.
- Import["file.bdf",{elem,suba,subb,…}] imports a subelement.
- Import["file.bdf",{{elem1,elem2,…}}] imports multiple elements.
- The import format can be specified with Import["file","BDF"] or Import["file",{"BDF",elem,…}].
- See the following reference pages for full general information:
-
Import import from a file CloudImport import from a cloud object ImportString import from a string ImportByteArray import from a byte array
Import Elements


- General Import elements:
-
"Elements" list of elements and options available in this file "Summary" summary of the file "Rules" list of rules for all available elements - Data representation elements:
-
"Data" all signal channels represented as a list of lists, with discontinuous records combined into a flat list {"Data",label} data from a named signal channel "Labels" names of all signal channels "LabeledData" list of rules, each representing a labeled data channel "RecordsData" all signal channels, given as an array of lists - Import uses the "Data" element by default.
- Import["file.bdf","Data"] gives a list of packed arrays of data samples, effectively combining discontinuous records for each signal channel into a flat list.
- Import["file.bdf","LabeledData"] imports the entire file and returns an expression of the form {label1->ch1,label2->ch2,…}.
- Import["file.bdf","RecordsData"] returns a packed array of the form {ch1,ch2,…}, where the signals chi are lists of discontinuous records {reci1,reci2,…}. Each record is given as a list of data samples.
- Metainformation elements:
-
"Annotations" time-stamped annotation list "ChannelCount" number of simultaneous signal channels "FilterInformation" information about signal filters applied during the recording, given as a list of strings "PatientID" patient identification string "Title" title of the recording, typically used as a document identification "RecordTimes" start times for each record, given in seconds "RecordCount" number of records in the file "RecordLength" length of records, given in seconds "DataRanges" physical data ranges for each signal channel "DataUnits" units in which the physical data for each channel is given "StartDate" local date and time when the recording was started "Device" information about the physical devices, such as electrodes, used for recording each channel - Import["file.bdf","Annotations"] gives the time-stamped annotation lists of a BDF file as a list of the form {{ti,duri}->{stri1,stri2,…},…}.
- BDF allows time gaps between records.
Examples
Basic Examples (3)
Show the labels of all signal channels available in a BDF file:
Import the labels and device names as a table:
Import the first record of the channel "A5" and plot it using ListLinePlot: