SFF (.sff)
- Import supports most common variants of the SFF file format, including those with and without an index.
Background & Context

-
- MIME type: chemical/seq-na-sff
- SFF molecular biology format.
- Standard flowgram format for storing and exchanging DNA sequences with base qualities.
- Commonly used by the 454 Life Sciences DNA pyrosequencing platform.
- Binary format.
- Stores nucleic acid sequences and base qualities as character strings and lists, respectively.
- Meta-information about the sequencing run are stored in the file.
Import

- Import["file.sff"] imports DNA sequencing data from an SFF file.
- Import["file.sff"] returns an array representing the sequencing data stored in the file.
- Import["file.sff",elem] imports the specified element from an SFF file.
- Import["file.sff",{{elem1,elem2,…}}] imports multiple elements.
- The import format can be specified with Import["file","SFF"] or Import["file",{"SFF",elem,…}].
- See the following reference pages for full general information:
-
Import import from a file CloudImport import from a cloud object ImportString import from a string ImportByteArray import from a byte array
Import Elements


- General Import elements:
-
"Elements" list of elements and options available in this file "Summary" summary of the file "Rules" list of rules for all available elements - File metadata:
-
"Header" file header given as a list of rules "XMLManifest" XML manifest as an XML object - Data representation elements for each sequencing read:
-
"Sequence" DNA sequences as a list of strings "Qualities" base qualities as a list of lists "FlowgramValues" flowgram values as a list of lists "FlowIndexPerBase" flow index values as a list of lists "ClipQualities" coordinates for quality-trimming the sequences as an array "ClipAdapter" coordinates for adapter-trimming the sequences as an array "ReadName" names of the reads as a list of strings - Additional data elements:
-
"Data" all data representation elements combined in a list "LabeledData" list of rules for each sequence stored in the file - Import uses the "Data" element by default for the SFF format.
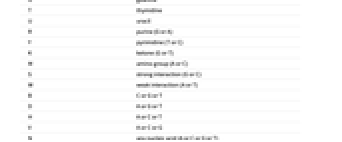
- The Wolfram Language uses the standard IUB/IUPAC abbreviations for nucleic acids:
-
A adenosine C cytidine G guanine T thymidine U uracil R purine (G or A) Y pyrimidine (T or C) K ketone (G or T) M amino group (A or C) S strong interaction (G or C) W weak interaction (A or T) B C or G or T D A or G or T H A or C or T V A or C or G N any nucleic acid (A or C or G or T) - gap of indeterminate length - The Wolfram Language uses integers for the base qualities.
Examples
open all close allRelated Guides
History
Introduced in 2012 (9.0)