ProteinData
ProteinData[entity]
gives the reference amino acid sequence for the protein entity.
ProteinData[entity,property]
gives the value of the specified property for the protein entity.
ProteinData[entity,property,annotation]
gives the specified annotation associated with the given property.
Details




- ProteinData[] gives a list of all reference human proteins.
- The specified entity in ProteinData can be an Entity, EntityClass, entity canonical name, or list thereof.
- The specified property can be an EntityProperty, EntityPropertyClass, property canonical name, or list of properties.
- Protein sequences are represented as strings of standard single-letter amino acid codes.
- Fundamental properties include:
-
"MolecularWeight" total molecular weight in daltons - Sequence properties for proteins include:
-
"DNACodingSequence" base pair sequence coding for the protein "DNACodingSequenceLength" length of base pair sequence coding for the protein "Gene" gene that codes for the protein "Sequence" amino acid sequence for the protein "SequenceLength" length of amino acid sequence for the protein - Protein structures may contain additional elements not explicitly encoded in the original DNA sequence.
- Molecular structure properties based on residues include:
-
"DihedralAngles" list of dihedral angles 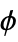 , ψ, ω in radians
, ψ, ω in radians"SecondaryStructureRules" list of rules giving start and end positions of helix, sheet, etc. structures - Molecular structure properties based on individual atoms include:
-
"AdditionalAtomPositions" list of 3D coordinates of additional atoms "AdditionalAtomTypes" list of element symbols for additional atoms "AtomPositions" list of 3D coordinates of protein atoms "AtomRoles" list of structural roles for protein atoms "AtomTypes" list of element symbols for protein atoms "GyrationRadius" radius of gyration "MoleculePlot" 3D molecular structure plot - Distances are measured in picometers.
- ProteinData[entity,property,grouping] gives molecular structure properties with various groupings:
-
{} no grouping "Chain" group by chain "Residue" group by residue {g1,g2,…} list of grouping criteria - Properties associated with chains within structures include:
-
"ChainLabels" list of identifiers for 3D structure chains "ChainSequences" list of amino acid sequences for 3D structure chains - Protein common domain properties include:
-
"DomainIDs" NCBI CDD numbers of domains "DomainPositions" positions of domains in the protein sequence "Domains" names of domains in the protein - Functional properties include:
-
"BiologicalProcesses" biological processes associated with the protein "CellularComponents" cellular components in which the protein is found "MolecularFunctions" molecular functions of the protein - Protein identification properties include:
-
"AlternateNames" alternate traditional names "GeneID" GeneID number string for the protein's gene "Name" traditional name "NCBIAccessions" NCBI accession strings "PDBIDList" list of all PDB ID strings "PrimaryPDBID" PDB ID chosen in the Wolfram Language for structure properties, etc. "StandardName" standard Wolfram Language name - ProteinData[entity,property,"Units"] gives the units for a particular property value.
Examples
open allclose allBasic Examples (6)
Scope (10)
Names and Classes (5)
Protein Structure (3)
Plot the ribbon diagram for a protein:
Get the 3D coordinates of each atom in a protein structure:
Get the corresponding atom types:
Group the atom coordinates by residue:
Group by chain and by residue:
Get the atom types for a particular residue:
Get the alpha-carbon atoms from each residue:
Use the alpha-carbon positions to render the protein backbone:
Properties & Relations (3)
Get the gene that encodes a protein:
Get the names of all proteins encoded by the gene from GenomeData:
Display a protein using atom sizes from ElementData and colorings from ColorData:
Show the conformation of a protein backbone using Tube and BezierCurve:
Text
Wolfram Research (2008), ProteinData, Wolfram Language function, https://reference.wolfram.com/language/ref/ProteinData.html (updated 2014).
CMS
Wolfram Language. 2008. "ProteinData." Wolfram Language & System Documentation Center. Wolfram Research. Last Modified 2014. https://reference.wolfram.com/language/ref/ProteinData.html.
APA
Wolfram Language. (2008). ProteinData. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/ProteinData.html