BioSequenceModify[seq,"mod"]
gives the result of applying the modification "mod" to the sequence seq.
BioSequenceModify[seq,{"mod",params}]
specifies the parameters params for "mod".
BioSequenceModify[modspec]
represents an operator form of BioSequenceModify that can be applied to a biomolecular sequence.




BioSequenceModify
BioSequenceModify[seq,"mod"]
gives the result of applying the modification "mod" to the sequence seq.
BioSequenceModify[seq,{"mod",params}]
specifies the parameters params for "mod".
BioSequenceModify[modspec]
represents an operator form of BioSequenceModify that can be applied to a biomolecular sequence.
Details

- Bond modifications:
-
{"AddBond",{i1,i2}} add a higher-order bond between letters at i1 and i2 {"AddBond",Bond[{i1,i2},"type"]} add a bond of the given type between the given indices {"DeleteBond",{i1,i2}} remove all higher-order bonds between the given indices {"DeleteBond",Bond[{i1,i2},"type"]} remove the specified bond between the given indices - Circularity adjustment modifications:
-
"MakeCircular" convert a linear sequence into a circular sequence "MakeLinear" convert a circular sequence into a linear sequence {"MakeLinear",i} convert to a linear sequence, starting at the i 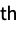 position
position - Collection modifications:
-
{"AddToCollection",{seq1,seq2,…}} incorporate a list of sequences into a sequence collection "SplitDisconnectedCollection" separate unbonded clusters into separate collections - Representation-only modifications:
-
"InnermostBondRepresentation" represent bonds at the innermost applicable sequence "OutermostBondRepresentation" represent bonds at the outermost sequence "CanonicalRepresentation" convert all sequences and bonds to a canonical form - Translation modifications:
-
"DropIncompleteCodons" drop incomplete codons from the end of DNA or RNA "DropToStartCodon" drop letters from DNA or RNA until a start codon is found "DropFromStopLetter" drop the letters from a peptide after a stop letter is found
Examples
open all close allBasic Examples (5)
Scope (30)
Convert a linear sequence to a circular sequence:
Convert a circular sequence to a linear sequence:
Add a list of sequences into a sequence collection:
Separate the unbound components of a sequence collection into separate collections:
Drop letters at the end of a nucleotide sequence so only complete codons are present for translation:
Drop the letters up to a start codon in the default genetic translation table:
Drop terms after the stop letter in a peptide sequence:
"AddBond" (4)
"AddToCollection" (3)
"CanonicalRepresentation" (3)
If sequences are identical, canonicalization will use strand-level bonds for ordering:
If the sequences and strand-level bonds are identical, canonicalization will use sequence bonds for ordering:
In addition to sorting, single-strand collections are reduced to the strand and single motif hybrids are reduced to the motif:
"DeleteBond" (2)
"DropToStartCodon" (3)
"InnermostBondRepresentation" (1)
"MakeCircular" (2)
"MakeLinear" (3)
"OutermostBondRepresentation" (1)
Applications (1)
Various peptides can be related to each other through a circular permutation:
Possible Issues (2)
Neat Examples (1)
Represent the protein preproinsulin as a BioSequence:
Remove the signal peptide sequence to make proinsulin:
Add the disulfide bonds and split the proinsulin sequence to make insulin:
See Also
BioSequence BioSequenceTranslate BioSequenceBackTranslateList
Entity Types: GeneticTranslationTable
Function Repository: BuildBioSequenceBondListFromFoldingMatrix
Related Guides
Text
Wolfram Research (2020), BioSequenceModify, Wolfram Language function, https://reference.wolfram.com/language/ref/BioSequenceModify.html (updated 2021).
CMS
Wolfram Language. 2020. "BioSequenceModify." Wolfram Language & System Documentation Center. Wolfram Research. Last Modified 2021. https://reference.wolfram.com/language/ref/BioSequenceModify.html.
APA
Wolfram Language. (2020). BioSequenceModify. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/BioSequenceModify.html
BibTeX
@misc{reference.wolfram_2025_biosequencemodify, author="Wolfram Research", title="{BioSequenceModify}", year="2021", howpublished="\url{https://reference.wolfram.com/language/ref/BioSequenceModify.html}", note=[Accessed: 27-February-2026]}
BibLaTeX
@online{reference.wolfram_2025_biosequencemodify, organization={Wolfram Research}, title={BioSequenceModify}, year={2021}, url={https://reference.wolfram.com/language/ref/BioSequenceModify.html}, note=[Accessed: 27-February-2026]}