WOLFRAM SYSTEM MODELER
SystemsBiologyFeatures for systems biology in SystemModeler |
|
Information
This document describes Systems Biology Extras for Wolfram SystemModeler™.
Introduction
Systems Biology Extras is an addition to SystemModeler and the BioChem library that provides a complete environment for modeling of biochemical systems. It adds libraries, tools, and features to SystemModeler in order to make modeling of such systems easier and more intuitive.
Enabling Systems Biology Extras
Systems Biology Extras is included in all SystemModeler licenses, but it is not enabled by default. To enable it, download and install the BioChem library from the WSM Library Store. Systems Biology Extras will enable itself when the BioChem library is loaded.
Creating Compartment Models
The compartment wizard of Systems Biology Extras lets you create compartment models in an intuitive step-by-step approach. A compartment model is a regular Modelica model that inherits specific interface models from the BioChem library.
You can create a compartment model by using the standard functionality provided in SystemModeler for creating classes, and later add other elements such as parameters and variables. This gives you the greatest level of flexibility and control. The compartment wizard, however, is designed specifically for the purpose of creating compartment models and has the advantage of being easier to use and lets you focus on the details of the compartment instead of Modelica language constructs. All compartment models created using the compartment wizard are also guaranteed to be exportable as SBML as long as they are not edited manually afterward.
To access the wizard, choose New Compartment Model from the Systems Biology menu. The compartment wizard consists of two dialog boxes. In the first dialog box you will be able to specify the name and other properties of the compartment model.
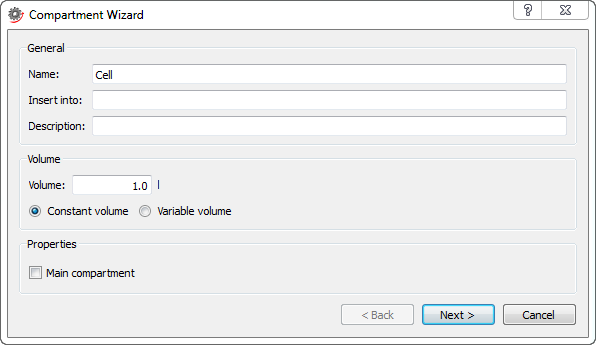
Specifying the properties of a compartment in the first step of the Compartment Wizard.
Name: the name of the compartment model.
Insert into (optional): a Modelica class path specifying where in the class hierarchy to create the compartment model. If no location is specified, the compartment model will be created at the top level of the class hierarchy.
Description (optional): describes the compartment model. The description appears along with the name of the model as balloon help for the model in the Class Browser.
Volume: the volume of the compartment. Select Constant volume to give the compartment a constant volume throughout the simulation. If you want the compartment volume to be able to vary throughout the simulation, select Variable volume.
Main compartment: this property is used to indicate that the compartment model is a main compartment. Checking this box is only relevant if your model has more than one compartment and you want to export the model to SBML.
Click the Next button to show the second and final dialog box that lets you add parameters and variables to the compartment model.
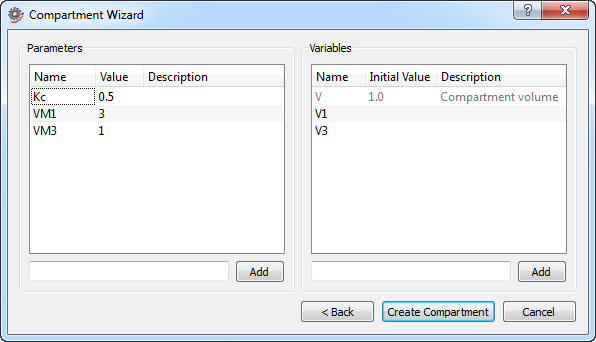
Adding parameters and variables in the second and final step of the Compartment Wizard.
To add a parameter to the compartment, enter the name of the parameter in the text box to the left and click the Add button (or press Enter). Variables are added in the same way by using the text box to the right. When adding a parameter or variable, its name appears in the list box. Double-click its name in the list box to rename it. It is also possible to give parameters and variables a value (initial value for variables) and a description by double-clicking in the corresponding column of the list box.
When all parameters and variables have been added, click the Create Compartment button to create the compartment model. The wizard will open the model and show it in the Diagram View automatically. The model will also be visible in the Class Browser at the location specified in the first step of the wizard.
Creating Reaction Models
Reaction models are most conveniently created using the reaction wizard, accessible from the Systems Biology menu. A reaction model is a regular Modelica model that inherits specific interface models from the BioChem library.
A reaction model can also be created using the standard functionality provided in SystemModeler for creating classes, in which you can later add elements such as a reaction rate formula and local parameters. This gives you the greatest level of flexibility and control. The reaction wizard, however, is designed for the sole purpose of creating reaction models and has the advantages of being more intuitive and easier to use, letting you focus on the details of the reaction instead of Modelica language constructs. All reaction models created using the reaction wizard are also guaranteed to fulfill the constraints that are required in order for a reaction model to be exportable as SBML. This is, of course, true only as long as the reaction model is not modified manually afterward in a way that breaks the constraints.
The reaction wizard is divided into two dialog boxes. In the first dialog you will be asked to specify the name and other basic attributes of the reaction.
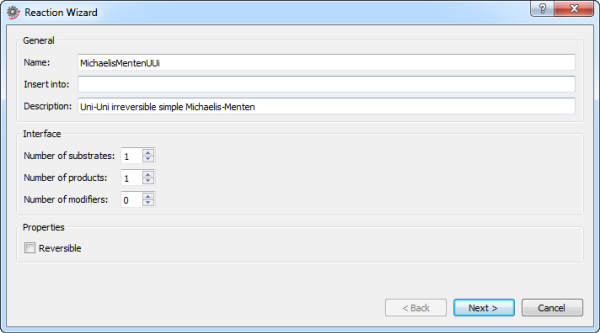
Specifying the attributes of a reaction in the first step of the Reaction Wizard.
Name: the name of the reaction model.
Insert into (optional): a Modelica class path specifying where in the class hierarchy to create the reaction model. If no location is specified, the reaction model will be created at the top level of the class hierarchy.
Description (optional): describes the reaction model. The description appears along with the name of the model as balloon help for the model in the Class Browser.
Number of substrates: specifies the number of substrate connectors in the reaction model.
Number of products: specifies the number of product connectors in the reaction model.
Number of modifiers: specifies the number of modifier connectors in the reaction model.
Reversible: specifies whether the reaction is reversible or irreversible.
Click the Next button to show the second and final dialog box of the reaction wizard. The final dialog box will let you create the parameters of the reaction and specify the reaction rate formula. The wizard provides buttons for built-in operators, functions, and constants as well as functions provided by the BioChem library. Any function can be referenced in the Reaction Rate Formula text box as long as you make sure it is available. If SBML exportability is important to you, we recommend that you read this chapter, describing the supported SBML subset, and more specifically the section on requirements that must to be fulfilled in order for a model to be exportable as SBML.
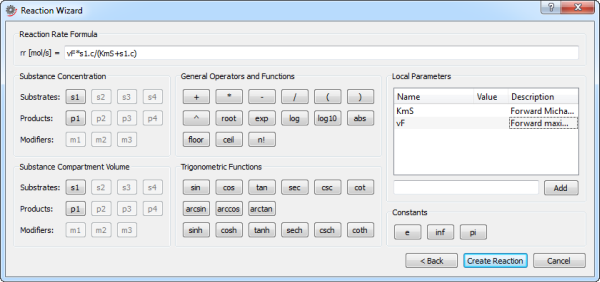
Specifying the reaction rate formula in the second and final step of the Reaction Wizard.
rr: the reaction rate formula specified in mol/s.
Substance Concentration: click the button of a substance to add a reference to its concentration in the reaction rate formula. The buttons represent the substance connected to the connector of the same name.
Substance Compartment Volume: click the button of a substance to add a reference to its compartment volume in the reaction rate formula. The buttons represent the substance connected to the connector of the same name.
General Operators and Functions: click the button of an operator or function to add it to the reaction rate formula.
Trigonometric Functions: click the button of a function to add it to the reaction rate formula.
Local Parameters: enter the name of the local parameter you wish to add to the reaction model and click the Add button (or press Enter). The parameter will appear in the list box. It is possible to rename a parameter as well as set its default value and a description by double-clicking in the corresponding column of the list box.
Constants: click the button of a constant to add it to the reaction rate formula.
Importing SBML Models
Models in the SBML format can be imported to Wolfram SystemModeler. When an SBML model is imported, it is automatically translated into Modelica code. To import an SBML model, choose File ▶ Import ▶ SBML and select the file that contains the SBML model.
The Messages view is updated with information related to the progress of the translation process. Check carefully for any warnings or errors once the translation of the model is complete. The most common warnings are related to unsupported elements and attributes of the SBML format.
If an SBML model fails the initial consistency checks that are applied before the actual translation of the model starts, the import will be aborted. A model failing the consistency checks is usually an indication of an incorrect SBML model. However, it may still be possible to import the model by inactivating one or more of the SBML consistency checks. For more information on how to change the settings of the SBML importer, see Settings below.
Settings
The consistency checks performed when importing an SBML model can be changed from the SBML Import view of the Options dialog box. Choose Tools ▶ Options to open the Options dialog box.
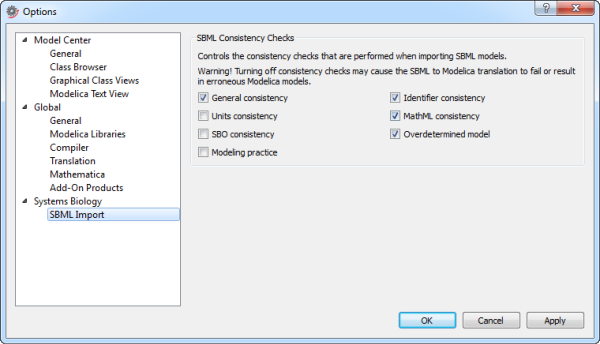
The SBML Import settings in the Options dialog box.
General consistency: correctness and consistency checks of specific SBML language constructs. Performing this set of checks is highly recommended.
Identifier consistency: correctness and consistency of identifiers used for model entities. An example of inconsistency would be using a species identifier in a reaction rate formula without first having declared the species.
Unit consistency: consistency checks of measurement units associated with quantities in a model.
MathML consistency: syntax check of MathML constructs.
SBO consistency: consistency and validity check of any Systems Biology Ontology (SBO) identifiers used in the model.
Overdetermined model: static analysis of whether the system of equations implied by a model is mathematically overdetermined.
Modeling practice: additional checks for recommended good modeling practice.
By default, the General consistency, Identifier consistency, MathML consistency, and Overdetermined model checks are enabled. These control points are highly recommended and any model following the SBML specification will pass these checks without warnings. Turning off one or more of these checks may cause the SBML-to-Modelica translation to fail, resulting in erroneous Modelica models.
Exporting SBML Models
Any model created with Systems Biology Extras that fulfills the constraints listed in this section can be exported to the SBML format. To export a model, right-click the main compartment model or the package containing the main compartment model in the Class Browser and choose Export ▶ SBML. The dialog box that appears will let you choose SBML version, SBML ID, and the SBML name of the model that will be created for your compartments.
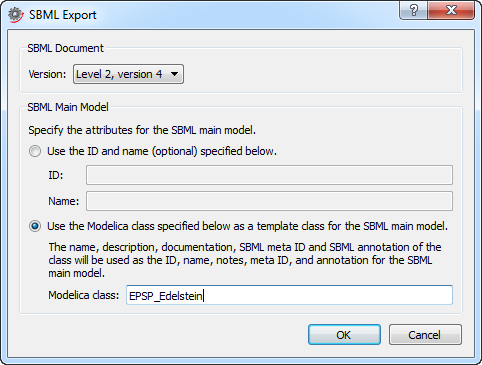
The SBML Export dialog box when exporting a package with a main compartment model.
The currently supported versions are level 2, version 1-4.
The SBML ID and name can either be set explicitly or by specifying the name of the corresponding Modelica class. If a Modelica class is specified, the ID, name, and notes of the SBML model will be extracted from the Modelica class. The class name becomes the SBML ID, the description the SBML name, and the documentation annotation the SBML notes.
In systems biology modeling in SystemModeler you will most likely create your main compartment model in a package and use the package as the template class when exporting your model to SBML. If you export a package containing a main compartment model, the name of the package will automatically appear in the Modelica class text box when the dialog box is shown.
Having selected a name for the SBML file after clicking the OK button of the dialog box, the translation of the model to SBML will begin. The Messages view is updated with information related to the progress of the translation process. Check carefully for any warnings or errors once the translation of the model is complete.
Metabolic Control Analysis
Metabolic Control Analysis (MCA) can be used to study the steady state properties of a model built with the BioChem package.
To perform an MCA, first simulate the model by pressing the Simulate Class button in the toolbar. In Simulation Center you can make any desired parameter adjustments in the Experiment Browser. The initial values that are set for the states are used as a starting guess for the steady state solver and may affect which solution the solver finds, so changing the initial values may be required. See the Simulation Center chapter in the SystemModeler User Guide for more information on how to change parameters and initial values.
Now choose Tools ▶ MCA. This will initiate a rebuild of the model with MCA capabilities enabled. During that process a perturbation parameter is added as a factor in all reaction rate equations. These parameters are later used to perturbate the reaction rates in the system. When the rebuild has finished, the MCA view is opened. Select the desired perturbation factor and solver tolerance and click Calculate. Now the steady state problem will be solved with one perturbated reaction rate at a time. The result is presented as normalized flux control coefficients and normalized concentration control coefficients. Positive coefficients are marked with a green color and negative coefficients with a red color.
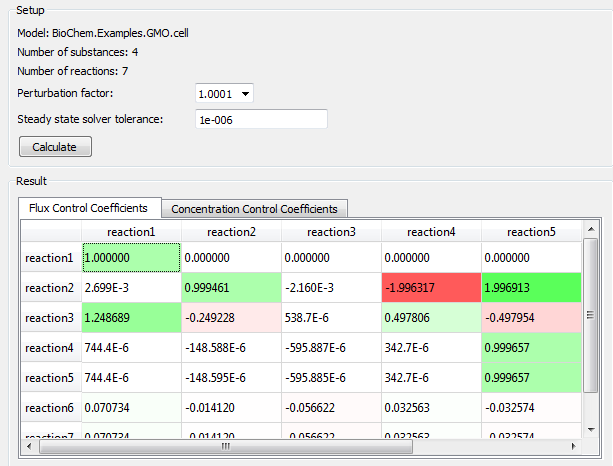
MCA view in Simulation Center.
Wolfram Language
SystemModel["BioChem.Documentation.SystemsBiology"]

