-
See Also
- Molecule
- BioSequence
- BioMoleculeValue
- BioMoleculePlot3D
- ExternalIdentifier
-
- Formats
- PDB
- MMCIF
-
- Service Connections
- ESMAtlas
- Related Guides
-
-
See Also
- Molecule
- BioSequence
- BioMoleculeValue
- BioMoleculePlot3D
- ExternalIdentifier
-
- Formats
- PDB
- MMCIF
-
- Service Connections
- ESMAtlas
- Related Guides
-
See Also
BioMolecule[id]
returns the biomolecule associated with the input identifier id.
BioMolecule[BioSequence["Peptide",seq]]
attempts to return a biomolecule representing a folded version of the input sequence seq.
BioMolecule[biomolpart]
returns a new biomolecule consisting of the part part of biomol.


BioMolecule
BioMolecule[id]
returns the biomolecule associated with the input identifier id.
BioMolecule[BioSequence["Peptide",seq]]
attempts to return a biomolecule representing a folded version of the input sequence seq.
BioMolecule[biomolpart]
returns a new biomolecule consisting of the part part of biomol.
Details and Options





- BioMolecule[input] is always converted to an optimized representation and treated as raw by functions like AtomQ and for purposes of pattern matching.
- In BioMolecule[id], id can be one of the following forms:
-
ExternalIdentifier[type,id] an external identifier BioSequence["Peptide",seq] a peptide sequence Entity["Protein",ent] a protein entity File[path] a "PDB" or "MMCIF" file - A BioMolecule consists of a collection of polymer chains, typically peptide, DNA or RNA, together with associated ligands and other nearby molecules.
- A BioMolecule always has coordinates, unlike BioSequence, which represents the primary structure of the biopolymer.
- A BioMolecule is a hierarchical object, with models containing chains containing residues containing atoms. Models represent different measurements of the same biomolecule and will vary in their 3D structure and possibly even atom counts. The majority of biomolecules from experimental structures contain just one model.
- Chains represent connected components of the biomolecule. Typically, each biopolymer is a separate chain, each ligand molecule is a chain, and any water molecules are grouped together in the last chain. Residues represent the individual monomers in a biopolymer chain and are made up of individual atoms with 3D coordinates.
- For inputs of the form BioMolecule[biomolpart], part should be an Association whose keys and default values are given by:
-
"Model" 1 which model to use "Chains" All which chains to include - BioMolecule[…][prop] returns a property of the biomolecule. Use BioMoleculeValue[biomolpart,prop] for property values specific to the given part of biomol.
- The following general properties are available:
-
"Name" biomolecule name "BioSequence" a BioSequence with type "HybridStrand" "ModelCount" number of models (usually one) "Models" a list of BioMolecule objects with one model each "AssemblyCount" number of assemblies "AssemblyData" data for assemblies "Assemblies" a list of BioMolecule objects with one assembly each "CrystallographicSpaceGroup" crystallographic space group "ComputedStructureQ" gives True if biomol is a predicted structure - Some biomolecules contain information about biological assemblies, which are the functional form of the molecule. These assemblies often differ from the asymmetric unit resolved from a crystal structure, often representing dimers, trimers or other complexes of the biomolecule chains.
- To create a biomolecule from a particular assembly, use BioMolecule[biomol<|"AssemblyNumber"n|>].
- The following properties apply at the model level:
-
"ChainCount" number of chains "ChainIDs" list of chain identifiers, e.g. {"A","B",…} "AuthorChainIDs" list of chain identifiers given by the original author "PolymerChainCount" number of polymer chains "PolymerChainLabels" list of labels for polymer chains "ResidueCount" number of residues in all chains - The following properties apply at the chain level, and the results are returned in an Association whose keys are the chain labels:
-
"ResidueCounts" number of residues per chain "ResidueAbbreviations" list of residue labels, e.g. {"Gly","Pro",…} "ResidueIDs" numeric identifiers "AuthorResidueIDs" numeric identifiers given by the original authors "AtomCounts" number of atoms per chain "ChainTypes" types of chains, e.g. {"Peptide","RNA",…} "BioSequences" BioSequence for each polymer chain "BackboneCoordinates" QuantityArray of backbone coordinates, "CA" for peptides and "O3" for nucleic acids "PeptideTorsionAngles" QuantityArray of peptide 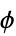 ,
, 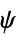 and
and 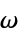 angles
angles"RamachandranAngles" QuantityArray of peptide 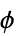 ,
, 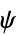 angles
angles"ResidueDistanceMatrix" distance matrix between backbone coordinates - The following properties apply at the residue level, and the results are returned in an Association whose keys are the chain labels and whose values are lists of lists, one for each residue:
-
"AtomicSymbols" symbols for the atoms "AtomCoordinates" atom coordinates with units, wrapped in QuantityArray "AtomCoordinateValues" numeric atom coordinates in angstroms "AtomLabels" labels for each atom, e.g. {"CA","CB",…} - BioMolecule[BioSequence[…]] will attempt to fold the given peptide sequence using the method specified by the option BioMoleculeFoldingMethod.
- Molecule[BioMolecule[…]] will return a Molecule object with bonds between individual atoms and hydrogen atoms added to fill in atomic valences.
Examples
open all close allBasic Examples (2)
Create a biomolecule from an ExternalIdentifier:
Scope (3)
Create a biomolecule from an ExternalIdentifier:
This biomolecule has two assemblies defined:
Retrieve the data about the assemblies:
Create a new biomolecule representing the second defined assembly:
View all the different models for a peptide:
Create a Molecule from a BioMolecule:
Possible Issues (1)
Some "Protein" entities cannot be converted to a BioMolecule:
See Also
Molecule BioSequence BioMoleculeValue BioMoleculePlot3D ExternalIdentifier
Service Connections: ESMAtlas
Related Guides
History
Text
Wolfram Research (2024), BioMolecule, Wolfram Language function, https://reference.wolfram.com/language/ref/BioMolecule.html.
CMS
Wolfram Language. 2024. "BioMolecule." Wolfram Language & System Documentation Center. Wolfram Research. https://reference.wolfram.com/language/ref/BioMolecule.html.
APA
Wolfram Language. (2024). BioMolecule. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/BioMolecule.html
BibTeX
@misc{reference.wolfram_2025_biomolecule, author="Wolfram Research", title="{BioMolecule}", year="2024", howpublished="\url{https://reference.wolfram.com/language/ref/BioMolecule.html}", note=[Accessed: 05-March-2026]}
BibLaTeX
@online{reference.wolfram_2025_biomolecule, organization={Wolfram Research}, title={BioMolecule}, year={2024}, url={https://reference.wolfram.com/language/ref/BioMolecule.html}, note=[Accessed: 05-March-2026]}