BioMoleculeValue[biomol,prop]
returns the value of the property prop for the BioMolecule biomol.
BioMoleculeValue[biomolpart,prop]
returns the property value association with part of biomol.


BioMoleculeValue
BioMoleculeValue[biomol,prop]
returns the value of the property prop for the BioMolecule biomol.
BioMoleculeValue[biomolpart,prop]
returns the property value association with part of biomol.
Details



- For inputs of the form BioMoleculeValue[biomolpart,prop], part should be an Association whose keys and default values are given by:
-
"Model" 1 which model to use "Chains" All which chains to include "Residues" All which residues to include "Atoms" All which atoms to include - The following general properties are available:
-
"Name" biomolecule name "BioSequence" a BioSequence with type "HybridStrand" "ModelCount" number of models (usually one) "Models" a list of BioMolecule objects with one model each "AssemblyCount" number of assemblies "AssemblyData" data for assemblies "Assemblies" a list of BioMolecule objects with one assembly each "CrystallographicSpaceGroup" crystallographic space group "ComputedStructureQ" gives True if biomol is a predicted structure "PDBString" a string representing biomol in the "PDB" format "MMCIFString" a string representing biomol in the "MMCIF" format - The following properties apply at the model level:
-
"ChainCount" number of chains "ChainIDs" list of chain identifiers, e.g. {"A","B",…} "AuthorChainIDs" list of chain identifiers given by the original author "PolymerChainCount" number of polymer chains "PolymerChainLabels" list of labels for polymer chains "ResidueCount" number of residues in all chains - The following properties apply at the chain level, and the results are returned in an Association whose keys are the chain labels:
-
"ResidueCounts" number of residues per chain "ResidueAbbreviations" list of residue labels, e.g. {"Gly","Pro",…} "ResidueIDs" numeric identifiers "AuthorResidueIDs" numeric identifiers given by the original authors "AtomCounts" number of atoms per chain "ChainTypes" types of chains, e.g. {"Peptide","RNA",…} "BioSequences" BioSequence for each polymer chain "BackboneCoordinates" QuantityArray of backbone coordinates, "CA" for peptides and "O3" for nucleic acids "PeptideTorsionAngles" QuantityArray of peptide 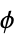 ,
, 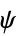 , and
, and 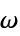 angles
angles"RamachandranAngles" QuantityArray of peptide 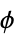 ,
, 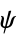 angles
angles"ResidueDistanceMatrix" distance matrix between backbone coordinates - The following properties apply at the residue level, and the results are returned in an Association whose keys are the chain labels and whose values are lists of lists, one for each residue:
-
"AtomicSymbols" symbols for the atoms "AtomCoordinates" atom coordinates with units, wrapped in QuantityArray "AtomCoordinateValues" numeric atom coordinates in angstroms "AtomLabels" labels for each atom, e.g. {"CA","CB",…}
Examples
open all close allBasic Examples (2)
Create a biomolecule from an ExternalIdentifier:
Create a biomolecule with multiple models:
Find the atom coordinate for the first atom in residue 3 for each model:
See Also
Related Guides
History
Text
Wolfram Research (2024), BioMoleculeValue, Wolfram Language function, https://reference.wolfram.com/language/ref/BioMoleculeValue.html.
CMS
Wolfram Language. 2024. "BioMoleculeValue." Wolfram Language & System Documentation Center. Wolfram Research. https://reference.wolfram.com/language/ref/BioMoleculeValue.html.
APA
Wolfram Language. (2024). BioMoleculeValue. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/BioMoleculeValue.html
BibTeX
@misc{reference.wolfram_2025_biomoleculevalue, author="Wolfram Research", title="{BioMoleculeValue}", year="2024", howpublished="\url{https://reference.wolfram.com/language/ref/BioMoleculeValue.html}", note=[Accessed: 19-February-2026]}
BibLaTeX
@online{reference.wolfram_2025_biomoleculevalue, organization={Wolfram Research}, title={BioMoleculeValue}, year={2024}, url={https://reference.wolfram.com/language/ref/BioMoleculeValue.html}, note=[Accessed: 19-February-2026]}