Simulation Center—Experiment Settings
| General | Input |
| Advanced | Output |
| Discretized Continuous-Time Partitions | Saving Experiment Settings in Model |
| Options |
The settings for a specific experiment can be changed from the Settings view of the Experiment Browser. Click the Settings tab to open the Settings view. It is also possible to set up default experiment settings that are used for all models that do not contain embedded experiment settings; see Options. The settings are divided into General, Advanced, Options, Input and Output.
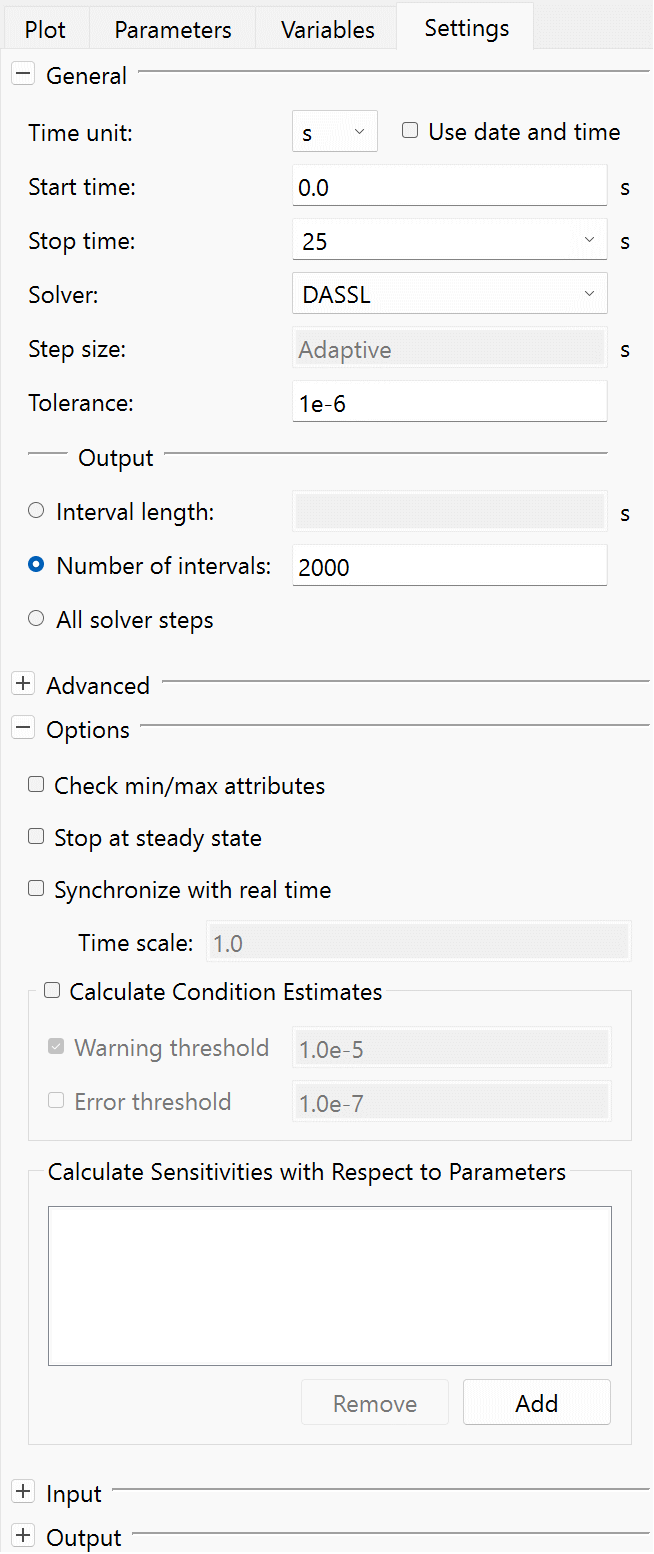
The Settings view of the Experiment Browser.
General
- Use date and time: toggles that the start and stop times of the simulation are specified as a date and time.
- Solver: Specifies the solver used to solve the dynamic system. There are five different solver choices:
- DASSL: a variable step size and variable order solver that uses a backward differentiation formula method.
- CVODES: A variable step size and variable order solver that uses a backward differentiation formula method. The added benefit of CVODES is that it supports forward sensitivity analysis. For more information on CVODES, see https://computing.llnl.gov/projects/sundials.
- Tolerance: Specifies the local tolerance that is used in each solver step. The final (global) error is in some way an accumulation of those per step errors. As a rule of thumb, reduce the desired global tolerance with a factor of 0.01 and use that as tolerance.
- Interval length: Specifies the length of the interval between output points from the solver. Note that the solver will output at least two data points for every event that is generated in the model in addition to what is specified by the interval length.
- Number of intervals: Specifies the number of output intervals that the solver generates. The default setting is 2000 intervals.
- All solver steps: When set, all internal solver steps are returned as result. This can potentially lead to huge datasets. Therefore, it might be better to use a fixed Number of intervals, or Interval length, for more complex models.
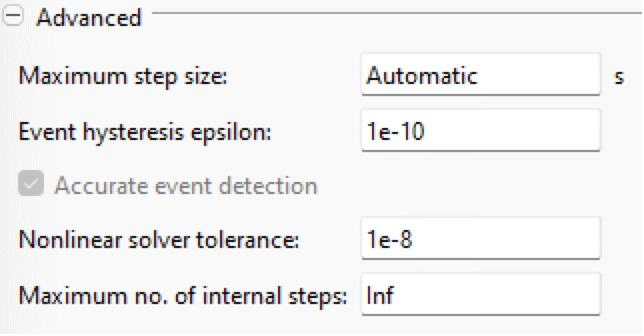
Advanced
- Simulation epoch: available if Use date and time is checked and specifies the date and time corresponding to time=0 in the simulation.
- Time zone: available if Use date and time is checked and specifies the time zone to use for setting and displaying date and time.
- Accurate event detection: Toggles root finding when using fixed-step solvers. If turned off, events will occur at the next output point.
- Maximum no. of internal steps: Specifies the maximum number of internal steps that the solver is allowed to take before reaching the next output point. This setting can be used to abort the simulation when the solver encounters a stiff system.
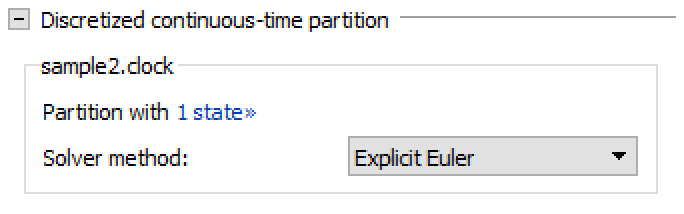
Discretized Continuous-Time Partitions
Settings for discretized continuous-time partitions.
Shows a list of all discretized continuous-time partitions with solverMethod="External". To view the states in a partition, click the number of states label. It is possible to select a specific solver method for each partition. There are three different solver methods to choose from:
The explicit method offers constant computation time, which can be desired in real-time scenarios. The implicit methods might need to be solved iteratively, so the computation time can vary; on the other hand, they offer increased stability that makes them more suitable for stiff systems.
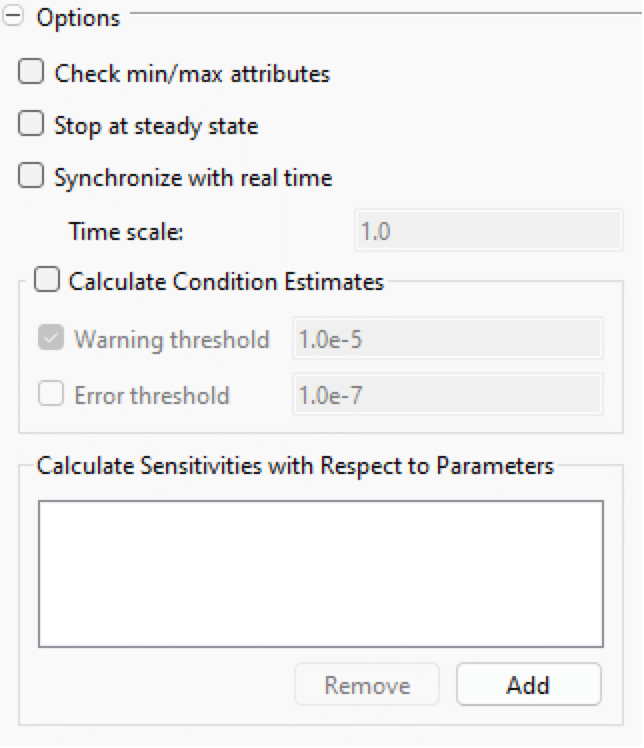
Options
- Check min/max attributes: if enabled, the simulation will terminate if any variable with min/max attributes goes outside its valid region.
- Stop at steady state: if enabled, the simulation will terminate if all state derivatives become equal to zero.
- Synchronize with real time: If enabled, the simulation will be synchronized with real time. That is, the simulation will be slowed down so that the simulation progresses with the same pace as real time. The real time will be scaled with the given Time scale; for example, a time scale of 2.0 would mean that the simulation progresses twice as fast as the wall clock time, so two simulated seconds will pass during one real second.
- Calculate Condition Estimates: if enabled, the simulation will estimate reciprocal condition numbers for all systems where that is applicable; for more information, see Condition Estimates.
- Calculate Sensitivities with Respect to Parameters: When the CVODES solver is used, it is possible to perform forward sensitivity analysis on parameters. To add a parameter for sensitivity analysis, click the Add button and select the desired parameter.
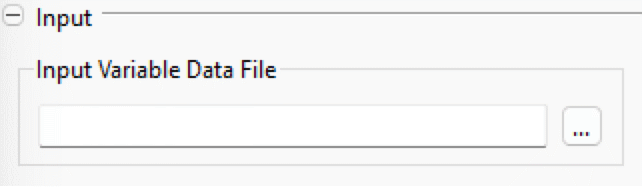
Input
- Input Variable Data File: specifies the trajectory for top-level inputs; see Input Variable Data File.
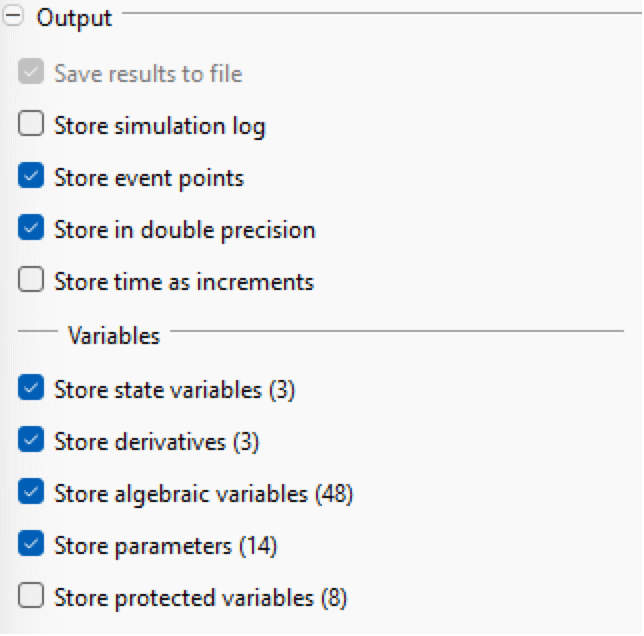
Output
- Save results to file: If enabled, the simulation result is written to a file. This is the default behavior and it only makes sense to uncheck this when running long real-time simulations.
- Store simulation log: if enabled, variables for the number of events, number of RHS evaluations and how real time progresses during the simulation are stored in the result file.
- Store event points: if enabled, the solver outputs extra output points at event points with the value before and after the event.
- Store in double precision: if enabled, the result is stored in double precision; otherwise, it is stored in single precision.
- Store time as increments: If enabled, the time vector is stored as time increments instead of the actual time values. This can eliminate precision problems for large simulation times when using single-precision storage.
- Store state variables: if enabled, all states (except protected if the store protected option is disabled) are stored in the result file.
- Store derivatives: if enabled, all derivatives (except protected if the store protected option is disabled) are stored in the result file.
- Store algebraic variables: if enabled, all algebraics (except protected if the store protected option is disabled) are stored in the result file.
- Store parameters: if enabled, all parameters (except protected if the store protected option is disabled) are stored in the result file.
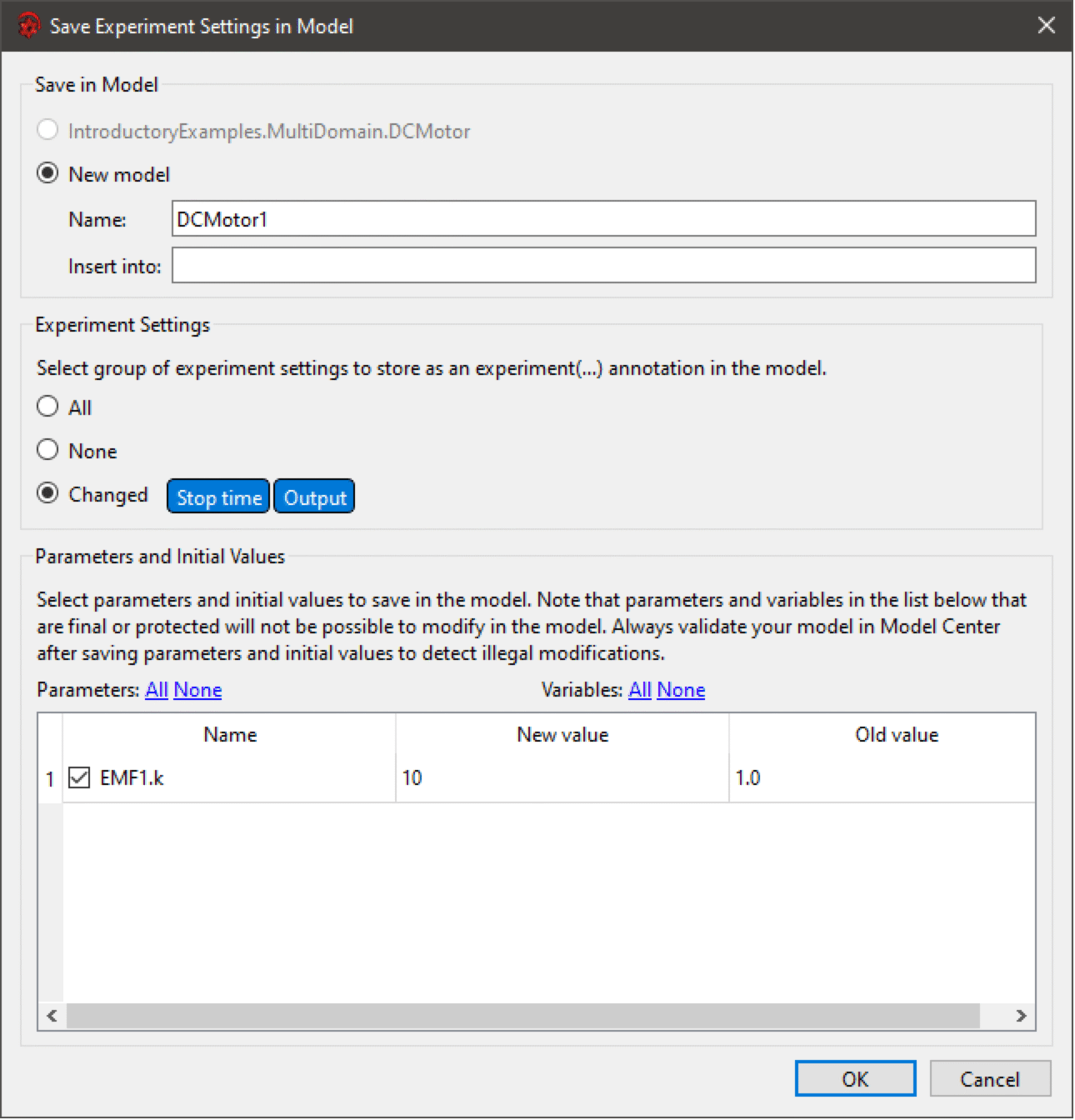
Saving Experiment Settings in Model
The Save Experiment Settings in Model dialog.
To save the experiment settings in the model, choose File ▶ Save Experiment Settings.
There are two ways of saving the experiment settings in the model:
Experiment settings, like time settings and solver settings, are saved using the experiment annotation. There are three possible choices for what to store:
- Changed: All changed settings will be saved in the experiment annotation. It is also possible to make a custom selection by clicking the individual settings tags.
Parameters and initial values are set directly or by using modifiers, depending on where in the hierarchy the variable is located.
After saving the settings, the model needs to be saved in Model Center for the changes to be permanent.