WOLFRAM SYSTEM MODELER
LibraryOverviewDescription of the library contents |
|
Information
The BioChem package was originally designed by Emma Larsdotter Nilsson and Erik Ulfheim. The library has since been further developed by Wolfram MathCore.
Introduction
When modeling biochemical reaction networks, the Systems Biology Markup Language (SBML) is often used. SBML has three main purposes:
- enabling the use of multiple software tools without having to rewrite the models for each tool
- enabling models to be shared and published in a form other researchers can use, even in a different software environment
- ensuring the survival of models beyond the lifetime of the software used to create them
However, the SBML language also has three major drawbacks:
- it is not possible to combine SBML models with models from other domains, such as electrical, mechanical, or hydraulic domains
- it is not convenient for modeling systems containing differential algebraic equations (DAE)
- though it is machine readable, it is not easily read by humans
By using Modelica and the BioChem library instead of SBML, these issues can be resolved while still meeting the original goal of SBML. Furthermore, the BioChem library has been developed in a way that facilitates the import and export of SBML models.
Library Content
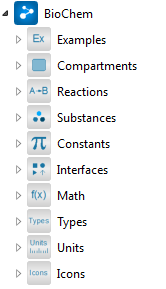
Packages in the BioChem library.
Since the design objective for BioChem is to provide properties and attributes that are common in biological and biochemical systems, the library contains several packages with standard components. Some of the packages consist of partial models, which are used as fundamental building blocks for complete models, while others contain components used for building up biochemical reactions.
The BioChem library consists of the following subpackages:
- Compartments - different types of compartments
- Constants - commonly used constants
- Examples - examples of pathway models
- Icons - the icons used in the library
- Interfaces - basic building blocks for biochemical components
- Math - mathematical expressions that are commonly used in SBML but are not included in the Modelica Standard Library
- Reactions - more than 200 different types of biochemical reactions
- Substances - pool of substances
- Units - units used in SBML that are not included in the Modelica Standard Library
- Types - package with BioChem specific types
The packages are shown in the figure above and are described in more detail in the following subsections.
Examples
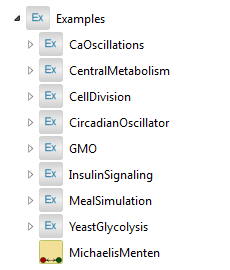
Contents of the Examples package.
The Examples package contains several model examples of pathways ranging from asymmetric reactions with Michaelis–Menten kinetics to more advanced multi-compartment models.
Compartments
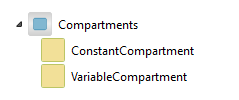
ConstantCompartment and VariableCompartment in the Compartments package.
This package contains compartment models. To control the environment of a chemical reaction during a simulation, it must take place in a restricted, screened-off container (a compartment). For this reason all models using the BioChem package must inherit a compartment.
The classes in the package are illustrated in the figure above. The difference between the two components is volume variability, which is either constant (ConstantCompartment) or varies over time (VariableCompartment). They both have an SBML-specific parameter, mainCompartment, that is used to indicate whether or not it is a main compartment. A main compartment is required when importing and exporting SBML models.
Reactions
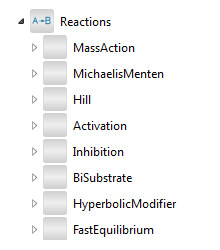
Contents of the Reactions package.
This package contains end-user models for a variety of reactions. All reactions have at least one substrate node and one product node, and in some cases they also have nodes for activators, inhibitors, or general modifiers. Each reaction is made up of the partial models in BioChem.Interfaces.Reactions.
The reaction models are divided into subpackages MassAction, MichaelisMenten, Hill, Activation, Inhibition, BiSubstrate, HyperbolicModifier, and FastEquilibrium.
MassAction
A sample of the reactions found in the MassAction package.
The mass action reactions are obtained by extending the reaction types in BioChem.Interfaces.Reactions and then adding an equation for the relation between the reaction rate and the participating substances, i.e. substrates, products, and modifiers. The mass action kinetics package is divided into the subpackages Irreversible and Reversible to facilitate navigation. These two packages contain the same reactions, the only difference being reversibility.
MichaelisMenten
Models in the MichaelisMenten package.
This package contains irreversible and reversible Michaelis–Menten reactions. Currently the library is limited to one-substrate, one-product reactions.
Michaelis–Menten kinetics describes the kinetics of many enzymes. It is relevant to situations where the concentration of enzyme is much lower than the concentration of substrate (i.e. where the enzyme concentration is the limiting factor) and when the enzyme is not allosteric.
Hill
Models in the Hill package.
In this package, models for Hill reaction kinetics with one or two modifiers are incorporated.
Activation
Models in the Activation package.
This package contains models for irreversible and reversible reactions where activators are involved.
Inhibition
Models in the Inhibition package.
In this package, different types of irreversible and reversible inhibition reaction kinetics are collected, e.g. competitive, non-competitive, and uncompetitive.
BiSubstrate
Models in the BiSubstrate package.
Ping pong and ordered reactions with one or two substrates or products are collected in this package.
HyperbolicModifier
Models in the HyperbolicModifier package.
This package contains models for irreversible and reversible hyperbolic modifier reaction kinetics.
FastEquilibrium
Models in the FastEquilibrium package.
The reactions in the FastEquilibrium package are used to model reactions that are very fast and that reach an almost instant state of balance. If translated to SBML, these models are approximated as very fast reactions.
Substances
Models in the Substances package.
The BioChem.Substances package contains different models needed to represent substances in biological and biochemical systems. The basic attributes corresponding to the properties that are studied during simulations (the amount n and the concentration c of the substance) are declared in these models.
The Substance model is used when the concentration in a substance node is allowed to change without restrictions during a simulation, while BoundarySubstance is used when the concentration can only be changed using events. This corresponds to species with the fixed or boundary attribute set in SBML.
The AmbientSubstance is a substance used as a reservoir in reactions. This corresponds to the empty list of substrates or the empty list of products in an SBML reaction.
When the concentration is not determined by reactions, the SignalSubstance model is used. Then the substance concentration is regulated by external equations, and it corresponds to SBML species changed by any SBML rules.
Constants
Contents of the Constants package.
The BioChem.Constants package contains mathematical constants that are needed for SBML import and export.
Interfaces

Contents of the Interfaces package.
The BioChem.Interfaces package contains partial models that can be used as building blocks for different types of nodes and reactions.
The models are designed to make it easy to create new customized components, as well as to make these components possible to translate to and from SBML.
The BioChem.Interfaces package contains the two subpackages Nodes and Reactions.
Nodes
Contents of the Nodes package.
The reaction between two or more substances is described partly by the molar flow rate between the involved substances. The molar flow rate is typically expressed as a function of the concentration or of the amount of substance. Therefore, the interfaces (nodes) contain not only the molar flow rate and concentration but also the volume, in order to allow the calculation of the amount of substance in moles.
The interfaces in the BioChem.Interfaces.Nodes library are all identical except for their graphics and the volume variable V, which is defined as either input or output. The different graphics are used to indicate if the substance should be seen as substrate, product, modifier, inhibitor, or an activator in a reaction.
| Variable Name | Description | Unit |
| c | concentration | mol l-1 |
| V | volume | l |
| r | reaction rate | mol/s |
Variables in BioChem.Interfaces.Nodes.
Reactions
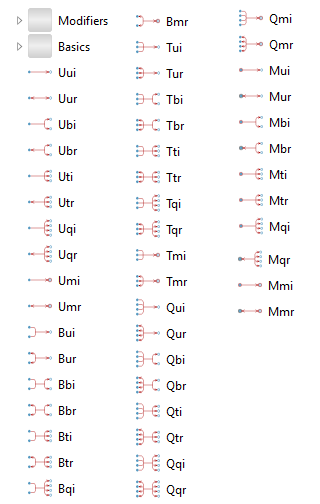
Building blocks for standard reactions.
The BioChem.Interfaces.Reactions package contains partial reaction models that can be used as templates when composing new reactions. In fact, all reactions found in BioChem.Reactions are based on these partial models.
All reactions need at least one substrate and one product. The BioChem.Interfaces.Reactions package contains base classes for any combination of substrates and products for reversible as well as irreversible reactions, as illustrated in the figure above.
The first letter in the reaction name indicates the number of substrates (see specification in the table below), and the second the number of products. Finally, the third letter indicates if the reaction is reversible (r) or irreversible (i). This information is also indicated by the icon.
| Abbreviation | Description |
| U | uni (one) |
| B | bi (two) |
| T | tri (three) |
| Q | quad (four) |
| M | multi (arbitrary) |
Abbreviation specification for the reaction in the BioChem.Interfaces package.
As an example, the following model extends the Bur (bi-uni-reversible) base model and has the addition of two parameters, k1 and k2, and an expression for the reaction rate.

Note that the equation for the reaction rate rr has been implemented using variables that are defined in the inherited base class BioChem.Interfaces.Reactions.Bur. The table below shows the variables that are available from the inherited class. Note that the available variables depend on the number of substrates and products in the inherited base class.
| Variable | Description |
| s1.c, s2.c | concentration of the first and second substrate |
| s1.V, s2.V | volume of the compartment in which the first and second substrate reside |
| s1.r, s2.r | total molar flow rate to the first and second substrate |
| p1.c | concentration of the first product |
| p1.V | volume of the compartment in which the first product resides |
| p1.r | total flow rate to the first product |
| nS1, nS2 | stoichiometric coefficient for the first and second substrate |
| nP1 | stoichiometric coefficient for the first product |
| rr | reaction rate |
Variables in the Bur model.
BioChem.Interfaces.Reactions also contains two subpackages, Modifiers and Basics.
Modifiers
Some reactions include one or more modifiers, activators, or inhibitors. In these cases, the partial models in the BioChem.Interfaces.Reactions.Modifiers should be used.
Basic building blocks to add modifiers to a reaction.
Creating reactions with modifiers is done by inheriting the corresponding modifier from the BioChem.Interfaces.Reactions.Modifiers package.
As an example, let us add an activator to a bi-uni reversible reaction (see below).

Note that we have added the variable a1.c to the equation for the reaction rate rr. This variable is accessible through the Activator model, where the connection point a1 is defined. The table below shows the variables that are available from the inherited BioChem.Interfaces.Reactions.Modifiers.Activator model.
| Variable | Description |
| a1.c | concentration of the first activator |
| a1.V | volume of the compartment in which the activator resides |
| a1.r | total flow rate to the first substrate |
Variables in the Activator model.
When a modifier is added, nodes (connection points) are added to the reaction according to the table below. Each node contains the variables for concentration, volume, and flow rate (c, V, r) for the respective modifier, as described in the section on nodes.
| Modifier Class | Name of Node |
| Activator | a1 |
| ActivatorBackward | aB1 |
| ActivatorForward | aF1 |
| LeftActivatorBackward | aB1 |
| LeftActivatorForward | aF1 |
| RightActivatorBackward | aB1 |
| RightActivatorForward | aF1 |
| Inhibitor | i1 |
| InhibitorBackward | iB1 |
| InhibitorForward | iF1 |
| LeftInhibitorBackward | iB1 |
| LeftInhibitorForward | iF1 |
| RightInhibitorBackward | iB1 |
| RightInhibitorForward | iF1 |
| ModifierBackward | mB1 |
| ModifierForward | mF1 |
| Modifier | m1 |
| TwoModifiers | m1, m2 |
| ThreeModifiers | m1, m2, m3 |
| MultipleModifiers | m1, m2,..., mn |
Modifier names.
Basics
Basic building blocks for reactions.
BioChem.Interfaces.Reactions.Basics contains the basic reaction models needed when defining a reaction.
The models available in this subpackage can be seen in the figure above. There are models for one to four substrates and products, as well as generic versions for multiple substrates and products. All substrate models come in two versions, one reversible variant and one irreversible. Finally, the Reaction class is the most basic class and is needed in all reactions.
When inheriting these classes, nodes (connection points) are added to the reaction according to the table below. Each node contains variables for concentration, volume, and flow rate (c, V, r).
| Model Name | Nodes (connectors) | Stoichiometric Coefficients |
| OneSubstrate OneSubstrateReversible | s1 | nS1 |
| TwoSubstrate TwoSubstratesReversible | s1, s2 | nS1, nS2 |
| ThreeSubstrates ThreeSubstratesReversible | s1, s2, s3 | nS1, nS2, nS3 |
| FourSubstrates FourSubstratesReversible | s1, s2, s3, s4 | nS1, nS2, nS3, nS4 |
| MultipleSubstrates | s1, s2,..., sn | nS1, nS2, ..., nSn |
| OneProduct | p1 | nP1 |
| TwoProducts | p1, p2 | nP1, nP2 |
| ThreeProducts | p1, p2, p3 | nP1, nP2, nP3 |
| FourProducts | p1, p2, p3, p4 | nP1, nP2, nP3, nP4s |
| MultipleProducts | p1, p2,..., pn | nP1, nP2, ..., nPn |
List of nodes and stoichiometric coefficients in models.
As an example, the BioChem.Interfaces.Reactions.Bui model is built up with models Reaction, TwoSubstrates, and Reversible, as shown below.

The BioChem.Interfaces.Reactions.Bui as seen in the text view.
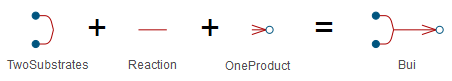
Illustration of inheritance for the BioChem.Interfaces.Reactions.Bui model.
Math

Functions in the Math package.
A number of mathematical functions are used in pathway models. Some of these can be found in Modelica.Math while others have been added to the Math package.
Types
The package BioChem.Types currently contains only one BioChem specific type, StateVariable. It is an enumeration type that allows the modeler to choose to use either the amount of substance n or the substance concentration c as a state.
Units
A number of physical types are needed to declare the different kinds of parameters and variables in the BioChem package. Some of the units of these types can be found in Modelica.Units.SI but they are here redefined to avoid long pathnames. The SI types used in BioChem are volume (l), amount of substance (mol), and concentration (mol m-3).
The package BioChem.Units also contains non-SI units, included in the table below, that are common in biochemical modeling.
| Quantity | Unit |
| AmountOfSubstance | mol |
| Concentration | mol l-1 |
| DilutionRate | s-1 |
| EquilibriumCoefficient | - |
| IonicCharge | - |
| MolarFlowRate | mol s-1 |
| Pressure | Pa |
| ReactionCoeff | - |
| ReactionOrder | - |
| ReactionRate | mol s-1 |
| StoichiometricCoefficient | - |
| Volume | l |
| VolumeChangeConstant | l s-1 |
| VolumetricReactionRate | mol l-1 s-1 |
The types and corresponding units used in BioChem.
Icons
The BioChem.Icons package contains icons that are used in the library.
Wolfram Language
SystemModel["BioChem.Documentation.LibraryOverview"]

