"SpanningTree" (Machine Learning Method)
- Method for FindClusters, ClusterClassify and ClusteringComponents.
- Partitions data into clusters of similar elements using the "SpanningTree" clustering algorithm.
Details & Suboptions
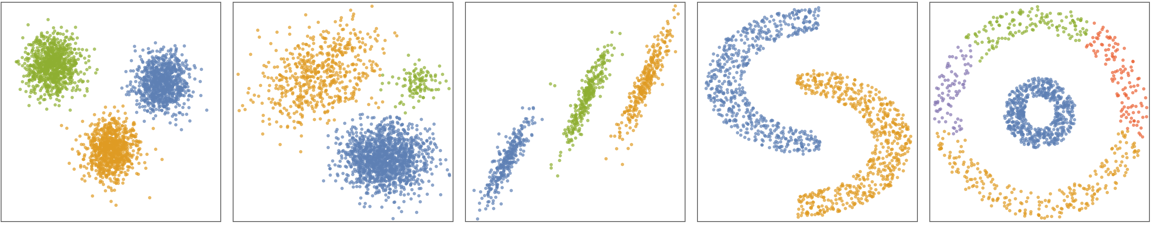
- "SpanningTree" is a neighbor-based clustering method. "SpanningTree" works for arbitrary cluster shapes and sizes; however, it can fail when clusters have different densities or are loosely connected.
- The following plots show the results of the "SpanningTree" method applied to toy datasets:
-
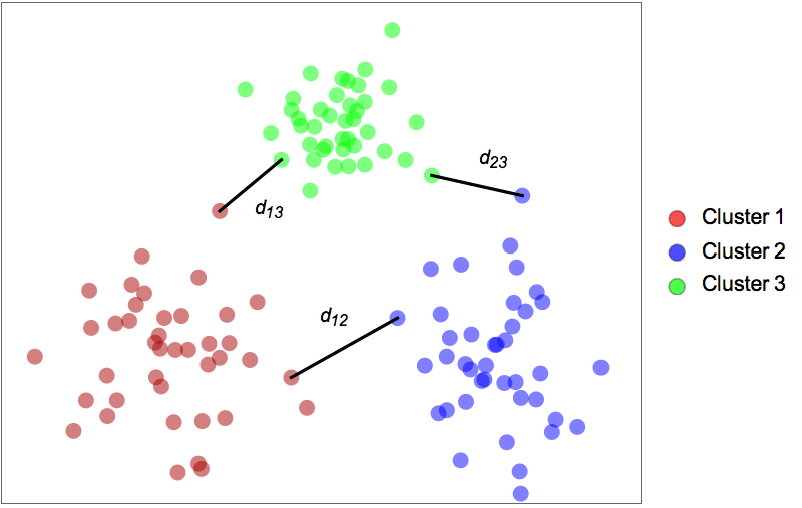
- The algorithm finds the set of clusters for which neighboring clusters are the most distant from each other. The distance dij between two neighboring clusters i and j is defined as the distance between their closest points:
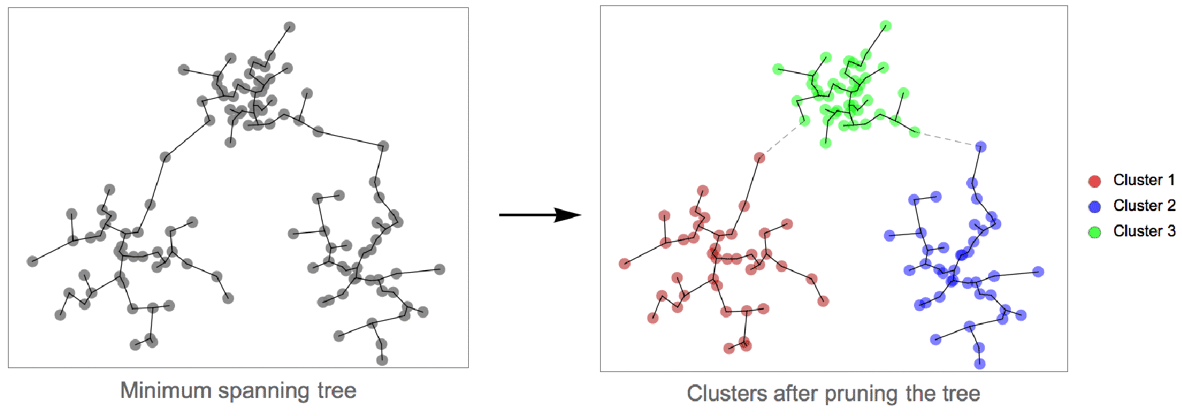
- Formally, the "SpanningTree" method constructs the minimum spanning tree of data points (using distances as graph weights). The longest edges of the tree are then pruned. Each connected component corresponds to a cluster. The pruning stops when the specified number of clusters is reached. When the number of clusters is not specified, the pruning stops when all edges are shorter than a given threshold.
- The option DistanceFunction can be used to define which distance to use.
- The following suboption can be given:
-
"MaxEdgeLength" Automatic pruning length threshold
Examples
open all close allBasic Examples (3)
Find clusters of nearby values using the "SpanningTree" method:
Train the ClassifierFunction using the "SpanningTree" method:
Scope (2)
Find cluster indices using ClusteringComponents:
Create and visualize noisy 2D moon-shaped training and test datasets:
Train a ClassifierFunction using "SpanningTree" and find clusters in the test set:
Visualizing two intertwined clusters found by "SpanningTree":