SmoothDensityHistogram[{{x1,y1},{x2,y2},…}]
plots a smooth kernel histogram of the values {xi,yi}.
SmoothDensityHistogram[{{x1,y1},{x2,y2},…},espec]
plots a smooth kernel histogram with estimator specification espec.
SmoothDensityHistogram[{{x1,y1},{x2,y2},…},espec,dfun]
plots the distribution function dfun.




SmoothDensityHistogram
SmoothDensityHistogram[{{x1,y1},{x2,y2},…}]
plots a smooth kernel histogram of the values {xi,yi}.
SmoothDensityHistogram[{{x1,y1},{x2,y2},…},espec]
plots a smooth kernel histogram with estimator specification espec.
SmoothDensityHistogram[{{x1,y1},{x2,y2},…},espec,dfun]
plots the distribution function dfun.
Details and Options


- SmoothDensityHistogram[data] by default generates colorized grayscale output of the PDF of {{x1,y1},…}, based on a smooth kernel density estimate.
- The estimator specification espec can be of the form bw or {bw,kernel}.
- The specifications for bandwidth bw and kernel are the same as for SmoothKernelDistribution.
- SmoothDensityHistogram[Tabular[…]cspec] extracts and plots values from the tabular object using the column specification cspec.
- The following forms of column specifications cspec are allowed for plotting tabular data:
-
{colx,coly} histogram values {x,y} from colx and coly - Possible distribution functions dfun include:
-
"PDF" probability density function "Intensity" count density function "CDF" cumulative distribution function "SF" survival function "HF" hazard function "CHF" cumulative hazard function - The form w[data] provides a wrapper w to be applied to the resulting graphics primitives.
- The following wrappers can be used:
-
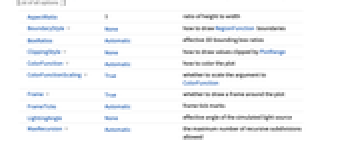
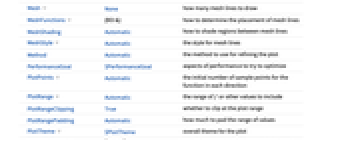
Annotation[e,label] provide an annotation Button[e,action] define an action to execute when the element is clicked EventHandler[e,…] define a general event handler for the element Hyperlink[e,uri] make the element act as a hyperlink PopupWindow[e,cont] attach a popup window to the element StatusArea[e,label] display in the status area when the element is moused over Style[e,opts] show the element using the specified styles Tooltip[e,label] attach an arbitrary tooltip to the element - SmoothDensityHistogram has the same options as Graphics, with the following additions and changes: [List of all options]
-
AspectRatio 1 ratio of height to width BoundaryStyle None how to draw RegionFunction boundaries BoxRatios Automatic effective 3D bounding box ratios ClippingStyle None how to draw values clipped by PlotRange ColorFunction Automatic how to color the plot ColorFunctionScaling True whether to scale the argument to ColorFunction Frame True whether to draw a frame around the plot FrameTicks Automatic frame tick marks LightingAngle None effective angle of the simulated light source MaxRecursion Automatic the maximum number of recursive subdivisions allowed Mesh None how many mesh lines to draw MeshFunctions {#3&} how to determine the placement of mesh lines MeshShading Automatic how to shade regions between mesh lines MeshStyle Automatic the style for mesh lines Method Automatic the method to use for refining the plot PerformanceGoal $PerformanceGoal aspects of performance to try to optimize PlotPoints Automatic the initial number of sample points for the function in each direction PlotRange Automatic the range of f or other values to include PlotRangeClipping True whether to clip at the plot range PlotRangePadding Automatic how much to pad the range of values PlotTheme $PlotTheme overall theme for the plot RegionFunction (True&) how to determine whether a point should be included ScalingFunctions None how to scale individual coordinates WorkingPrecision MachinePrecision the precision used in internal computations - ColorFunction is supplied with a single argument, given by default by the scaled value of f, the PDF, CDF, etc. of the distribution.
- The arguments supplied to RegionFunction are x, y, and f, where f can be the PDF, CDF, etc. of the distribution.
- With the setting MeshShading->Automatic, the colors between mesh lines are computed from the ColorFunction setting.
- With ScalingFunctions->{sx,sy,sz}, the
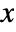 coordinate is scaled using sx, the
coordinate is scaled using sx, the 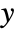 coordinate is scaled using sy, and the
coordinate is scaled using sy, and the 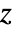 coordinate is scaled using sz.
coordinate is scaled using sz. -
 Highlight options with settings specific to SmoothDensityHistogram
Highlight options with settings specific to SmoothDensityHistogram
-
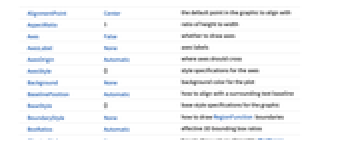
AlignmentPoint Center the default point in the graphic to align with AspectRatio 1 ratio of height to width Axes False whether to draw axes AxesLabel None axes labels AxesOrigin Automatic where axes should cross AxesStyle {} style specifications for the axes Background None background color for the plot BaselinePosition Automatic how to align with a surrounding text baseline BaseStyle {} base style specifications for the graphic BoundaryStyle None how to draw RegionFunction boundaries BoxRatios Automatic effective 3D bounding box ratios ClippingStyle None how to draw values clipped by PlotRange ColorFunction Automatic how to color the plot ColorFunctionScaling True whether to scale the argument to ColorFunction ContentSelectable Automatic whether to allow contents to be selected CoordinatesToolOptions Automatic detailed behavior of the coordinates tool Epilog {} primitives rendered after the main plot FormatType TraditionalForm the default format type for text Frame True whether to draw a frame around the plot FrameLabel None frame labels FrameStyle {} style specifications for the frame FrameTicks Automatic frame tick marks FrameTicksStyle {} style specifications for frame ticks GridLines None grid lines to draw GridLinesStyle {} style specifications for grid lines ImageMargins 0. the margins to leave around the graphic ImagePadding All what extra padding to allow for labels etc. ImageSize Automatic the absolute size at which to render the graphic LabelStyle {} style specifications for labels LightingAngle None effective angle of the simulated light source MaxRecursion Automatic the maximum number of recursive subdivisions allowed Mesh None how many mesh lines to draw MeshFunctions {#3&} how to determine the placement of mesh lines MeshShading Automatic how to shade regions between mesh lines MeshStyle Automatic the style for mesh lines Method Automatic the method to use for refining the plot PerformanceGoal $PerformanceGoal aspects of performance to try to optimize PlotLabel None an overall label for the plot PlotPoints Automatic the initial number of sample points for the function in each direction PlotRange Automatic the range of f or other values to include PlotRangeClipping True whether to clip at the plot range PlotRangePadding Automatic how much to pad the range of values PlotRegion Automatic the final display region to be filled PlotTheme $PlotTheme overall theme for the plot PreserveImageOptions Automatic whether to preserve image options when displaying new versions of the same graphic Prolog {} primitives rendered before the main plot RegionFunction (True&) how to determine whether a point should be included RotateLabel True whether to rotate y labels on the frame ScalingFunctions None how to scale individual coordinates Ticks Automatic axes ticks TicksStyle {} style specifications for axes ticks WorkingPrecision MachinePrecision the precision used in internal computations
List of all options
Examples
open all close allBasic Examples (2)
Scope (23)
Data and Wrappers (8)
Tabular Data (1)
Bandwidth and Kernel (9)
Specify a single bandwidth for bivariate data:
Specify bivariate bandwidths in units of standard deviation:
Allow bivariate bandwidths to vary adaptively with local density:
Use the local sensitivity from ![]() (small) to
(small) to ![]() (large):
(large):
Vary the initial bandwidth for an adaptive estimate:
Use initial bandwidths of ![]() and
and ![]() :
:
Use any of several automatic bandwidth selection methods:
Silverman's method is used by default for bandwidth selection:
Use different bandwidth specifications in each dimension:
Options (68)
AspectRatio (4)
By default, SmoothDensityHistogram uses the same width and height:
Use a numerical value to specify the height to width ratio:
AspectRatioAutomatic determines the ratio from the plot ranges:
AspectRatioFull adjusts the height and width to tightly fit inside other constructs:
Axes (3)
By default, SmoothDensityHistogram uses a frame instead of axes:
AxesLabel (4)
AxesOrigin (2)
AxesStyle (3)
BoundaryStyle (2)
Use a red boundary around the edges of the surface:
BoundaryStyle applies to regions cut by RegionFunction:
ClippingStyle (4)
ColorFunction (5)
ColorFunctionScaling (1)
Color using the natural range of values by setting ColorFunctionScaling to False:
ImageSize (7)
Use named sizes such as Tiny, Small, Medium and Large:
Specify the width of the plot:
Specify the height of the plot:
Allow the width and height to be up to a certain size:
Specify the width and height for a graphic, padding with space if necessary:
Setting AspectRatioFull will fill the available space:
Use maximum sizes for the width and height:
Use ImageSizeFull to fill the available space in an object:
Specify the image size as a fraction of the available space:
Mesh (7)
SmoothDensityHistogram typically has 10 mesh lines in the ![]() direction:
direction:
Use 5 mesh lines in the ![]() direction:
direction:
Show the complete sampling mesh:
Use 3 mesh lines in the ![]() direction and 6 mesh lines in the
direction and 6 mesh lines in the ![]() direction:
direction:
MeshFunctions (3)
MeshStyle (2)
PlotRange (3)
SmoothHistogram3D automatically selects the domain:
Use the full domain generated by SmoothKernelDistribution:
PlotTheme (2)
RegionFunction (4)
Clip small values of the surface:
BoundaryStyle is used where the region is clipped:
Ticks (4)
Applications (2)
Properties & Relations (7)
SmoothDensityHistogram effectively plots the distribution function of SmoothKernelDistribution:
Use DensityHistogram to plot the data in discrete bins:
Use SmoothDensityHistogram and SmoothHistogram3D for bivariate data:
Use SmoothHistogram for univariate data:
Use GeoSmoothHistogram for geographic data:
Additional points will result in a better approximation of the underlying distribution:
As the bandwidth approaches infinity, the estimate approaches the shape of the kernel:
Related Guides
History
Introduced in 2010 (8.0) | Updated in 2014 (10.0) ▪ 2015 (10.2) ▪ 2025 (14.2)
Text
Wolfram Research (2010), SmoothDensityHistogram, Wolfram Language function, https://reference.wolfram.com/language/ref/SmoothDensityHistogram.html (updated 2025).
CMS
Wolfram Language. 2010. "SmoothDensityHistogram." Wolfram Language & System Documentation Center. Wolfram Research. Last Modified 2025. https://reference.wolfram.com/language/ref/SmoothDensityHistogram.html.
APA
Wolfram Language. (2010). SmoothDensityHistogram. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/SmoothDensityHistogram.html
BibTeX
@misc{reference.wolfram_2025_smoothdensityhistogram, author="Wolfram Research", title="{SmoothDensityHistogram}", year="2025", howpublished="\url{https://reference.wolfram.com/language/ref/SmoothDensityHistogram.html}", note=[Accessed: 21-February-2026]}
BibLaTeX
@online{reference.wolfram_2025_smoothdensityhistogram, organization={Wolfram Research}, title={SmoothDensityHistogram}, year={2025}, url={https://reference.wolfram.com/language/ref/SmoothDensityHistogram.html}, note=[Accessed: 21-February-2026]}