MoleculeValuePlot[mol,"prop"]
returns a two-dimensional plot of the molecule mol with colors corresponding to the atom or bond property "prop".
MoleculeValuePlot[mol,{item1val1,…}]
returns a molecule plot with atoms or bonds colored according to the specified values.
MoleculeValuePlot[mol,func]
uses the pure function func to determine values by which to color atoms or bonds.


MoleculeValuePlot
MoleculeValuePlot[mol,"prop"]
returns a two-dimensional plot of the molecule mol with colors corresponding to the atom or bond property "prop".
MoleculeValuePlot[mol,{item1val1,…}]
returns a molecule plot with atoms or bonds colored according to the specified values.
MoleculeValuePlot[mol,func]
uses the pure function func to determine values by which to color atoms or bonds.
Details



- Get a molecule plot with atoms or bonds colored according to property values
- MoleculeValuePlot uses color to associate numeric or categorical values with regions in a molecule.
- mol can be a Molecule object or something that can be easily converted to one, such as a systematic chemical name, a "Chemical" entity, a BioSequence or an ExternalIdentifier.
- "prop" should be a Molecule property that returns values for atoms or bonds.
- func should be a pure function using named slots, e.g. (#AtomicNumber/#FormalCharge&).
- EntityProperty["Element",…] can be used to plot the property values for the types of elements in the molecule.
- Use MoleculeValue["AtomProperties"] for a full list of per-atom properties. The following list includes typical atom properties:
-
"AromaticAtomQ" whether the atom is aromatic "AtomChirality" absolute atomic chirality, e.g. R, S, for chiral atoms "AtomicMass" atomic mass "AtomicNumber" atomic number "AtomicSymbol" IUPAC atomic symbol "AtomSMARTS" atom SMARTS "AtomSMILES" atom SMILES "CoordinationNumber" number of explicit bonds for an atom "FormalCharge" formal charge "GasteigerPartialCharge" atomic charge using the Gasteiger charge model "GeometricStericEffectIndex" geometric steric index; requires atomic coordinates "HydrogenCount" number of hydrogens bonded to the atom "MMFFPartialCharge" atomic charge computed using the MMFF force field "OrbitalHybridization" computed orbital hybridization "OuterShellElectronCount" number of outer-shell electrons "OxidationState" the degree of oxidation of an atom in a molecule "PiElectronCount" number of 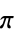 electrons
electrons"TopologicalStericEffectIndex" topological steric effect index "UnpairedElectronCount" number of unpaired electrons - Use MoleculeValue["BondProperties"] for a full list of per-bond properties. The following list includes typical bond properties:
-
"BondIndex" position of a bond in the bond list for a molecule "BondLength" Euclidean distance between the bonded atoms "BondOrder" number of bonding electron pairs "BondStereo" absolute bond stereo, e.g. E or Z "BondType" bond type, e.g. Single, Double, etc. "ConjugatedBondQ" whether the bond is part of a conjugated system - MoleculeValuePlot has the same options as Graphics, with the following additions and changes: [List of all options]
-
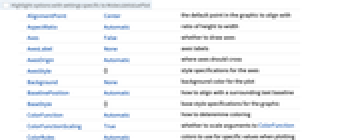
ColorFunction Automatic how to deteremine coloring ColorFunctionScaling True whether to scale arguments to ColorFunction ColorRules Automatic colors to use for specific values when plotting categorical data IncludeHydrogens Automatic whether to show hydrogen atoms PlotLegends Automatic legends for highlights PlotTheme $PlotTheme overall theme for the plot Method Automatic can be "Numerical" or "Categorical" -
AlignmentPoint Center the default point in the graphic to align with AspectRatio Automatic ratio of height to width Axes False whether to draw axes AxesLabel None axes labels AxesOrigin Automatic where axes should cross AxesStyle {} style specifications for the axes Background None background color for the plot BaselinePosition Automatic how to align with a surrounding text baseline BaseStyle {} base style specifications for the graphic ColorFunction Automatic how to deteremine coloring ColorFunctionScaling True whether to scale arguments to ColorFunction ColorRules Automatic colors to use for specific values when plotting categorical data ContentSelectable Automatic whether to allow contents to be selected CoordinatesToolOptions Automatic detailed behavior of the coordinates tool Epilog {} primitives rendered after the main plot FormatType TraditionalForm the default format type for text Frame False whether to put a frame around the plot FrameLabel None frame labels FrameStyle {} style specifications for the frame FrameTicks Automatic frame ticks FrameTicksStyle {} style specifications for frame ticks GridLines None grid lines to draw GridLinesStyle {} style specifications for grid lines ImageMargins 0. the margins to leave around the graphic ImagePadding All what extra padding to allow for labels etc. ImageSize Automatic the absolute size at which to render the graphic IncludeHydrogens Automatic whether to show hydrogen atoms LabelStyle {} style specifications for labels Method Automatic can be "Numerical" or "Categorical" PlotLabel None an overall label for the plot PlotLegends Automatic legends for highlights PlotRange All range of values to include PlotRangeClipping False whether to clip at the plot range PlotRangePadding Automatic how much to pad the range of values PlotRegion Automatic the final display region to be filled PlotTheme $PlotTheme overall theme for the plot PreserveImageOptions Automatic whether to preserve image options when displaying new versions of the same graphic Prolog {} primitives rendered before the main plot RotateLabel True whether to rotate y labels on the frame Ticks Automatic axes ticks TicksStyle {} style specifications for axes ticks
List of all options


Examples
open all close allBasic Examples (4)
Scope (2)
Options (4)
Method (1)
By default, numeric data is plotted using a continuous color function:
Use Method"Categorical" to force a different legend type:
PlotLegends (1)
Plot legends are shown by default for most inputs:
Use PlotLegendsNone to disable automatic legends:
Specify a specific legend placement and options for the legend:
See Also
History
Text
Wolfram Research (2025), MoleculeValuePlot, Wolfram Language function, https://reference.wolfram.com/language/ref/MoleculeValuePlot.html.
CMS
Wolfram Language. 2025. "MoleculeValuePlot." Wolfram Language & System Documentation Center. Wolfram Research. https://reference.wolfram.com/language/ref/MoleculeValuePlot.html.
APA
Wolfram Language. (2025). MoleculeValuePlot. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/MoleculeValuePlot.html
BibTeX
@misc{reference.wolfram_2025_moleculevalueplot, author="Wolfram Research", title="{MoleculeValuePlot}", year="2025", howpublished="\url{https://reference.wolfram.com/language/ref/MoleculeValuePlot.html}", note=[Accessed: 28-February-2026]}
BibLaTeX
@online{reference.wolfram_2025_moleculevalueplot, organization={Wolfram Research}, title={MoleculeValuePlot}, year={2025}, url={https://reference.wolfram.com/language/ref/MoleculeValuePlot.html}, note=[Accessed: 28-February-2026]}